Targeting Individual Cells to Fight Antimicrobial Resistance
New molecular tools reveal diversity in bacterial communities.
Treatment for a bacterial infection has two basic steps: Identify the species of bacteria; then administer an antibiotic that kills that species. But increasingly, antimicrobial resistance threatens the effectiveness of established regimens.
One solution may lie in bacterial communities themselves—which are more diverse than previously believed, says Kimberly Davis, PhD, MSc, an assistant professor in Molecular Microbiology and Immunology. Subpopulations of bacteria may differ considerably in growth rate or response to drugs, for example. Exploiting those differences could be key to fighting infections.
Scientists only recently acquired the means to observe such bacterial diversity. “New single-cell techniques … allow you to isolate individual bacteria and study them in more detail, including single-cell sequencing and fluorescence-activated cell sorting,” Davis says.
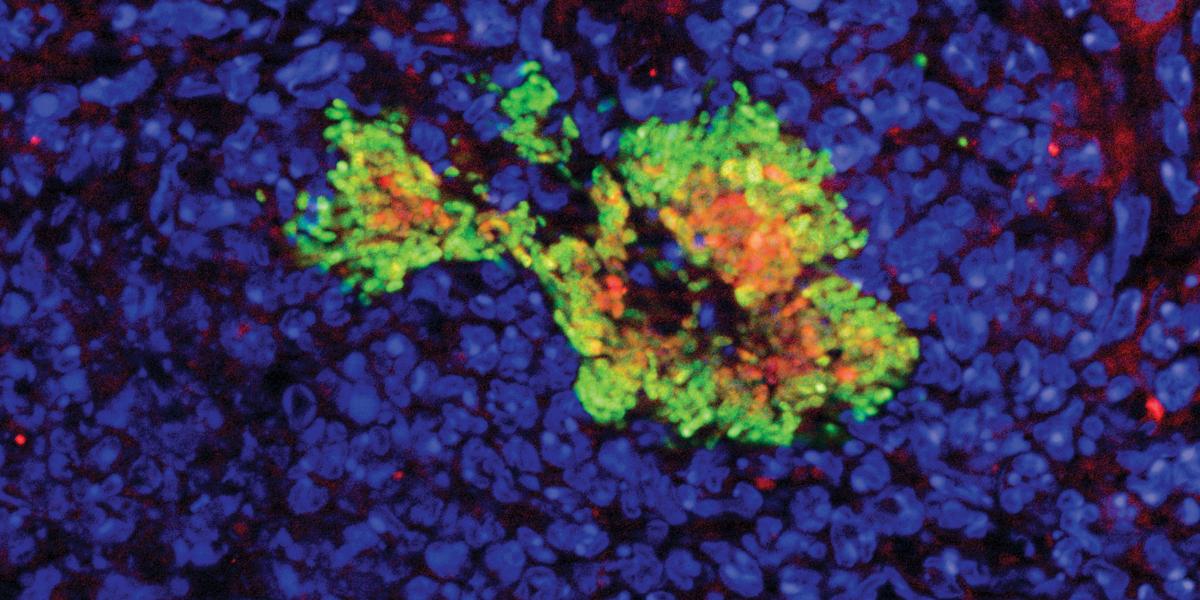
During her postdoctoral research, using the latter technique to study Yersinia pseudotuberculosis, Davis showed that the host immune response has differential effects on subpopulations of bacteria. For example, some immune cells secrete antimicrobial compounds that affect bacterial cells they contact directly—those on an infection’s periphery—but not the cells deep within. “Direct contact with different immune cells can alter the gene expression of the bacteria,” she says.
To flag individual cells’ responses and behaviors, Davis’ lab has developed several fluorescent “reporters”—DNA sequences engineered to generate proteins that glow when certain genes are turned on, or in response to specific biological events. For example, some reporters light up when the rate of cell division accelerates or slows down, or report how bacteria are responding to treatment with antibiotics.
The resulting insights into the dynamics of bacterial communities could reveal previously overlooked vulnerabilities or lead to more individualized care for patients with infections. Davis is especially keen to apply these tools to Staphylococcus aureus, a dangerous bacterium that can acquire broad resistance to existing antibiotics. “We need to start to think about alternatives [to existing antibiotics],” she says, “whether it be combinations of therapeutics or targeting specific proteins present in that particular infection.”