Survival Through Difference: How Bacteria Resist Treatment
New tools reveal how differences in individual cells within bacterial communities help them survive—and suggest targets for overcoming drug resistance.
A football game can be hopelessly confusing to a non-fan, with lines of identically clad players colliding into a jumbled mass. But experienced spectators see individual players and can discriminate each team’s strategies for seizing victory.
Kimberly Davis, PhD, MSc, aims to apply a similar tactical lens to bacterial infections. Her team is developing tools and methods for monitoring what’s happening in individual cells within bacterial communities as they defend themselves—both against the host immune system and antibiotic drugs.
Understanding this hidden heterogeneity is critical for the development of more effective treatments that minimize the risk of antibiotic resistance. “Oftentimes when a patient takes antibiotics, the vast majority of the [bacterial] population may respond quite well, but not all of the population will,” says Brian Conlon, PhD, a colleague of Davis’ who studies antibiotic resistance at the University of North Carolina. “And in that scenario, we need to know about those really problematic subpopulations that don't respond to drugs.”
Davis’ experimental bacterium of choice is Yersinia pseudotuberculosis, a direct ancestor of Yersinia pestis—the pathogen responsible for plague, which caused the Black Death. Although less dangerous than its descendant, Y. pseudotuberculosis can still cause serious disease in both humans and mice, making it ideal for modeling and studying the course of clinical disease in the lab.
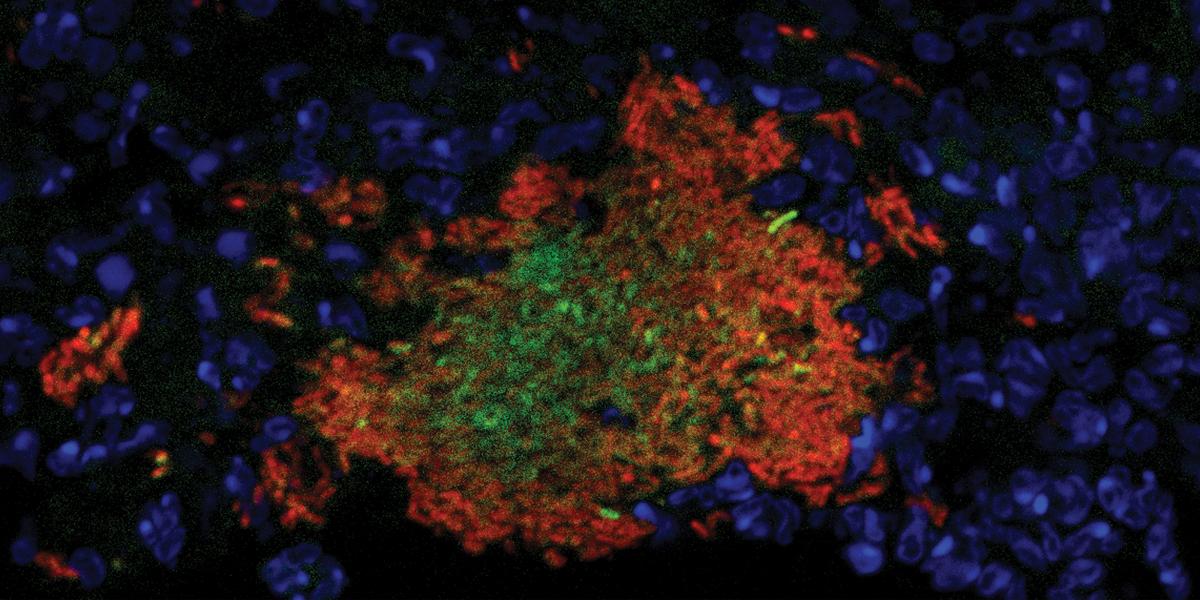
During infection, these bacteria form compact clusters. The host’s immune system soon surrounds them with cells called neutrophils. As the community fends off the immune system’s attempts to eradicate it, clear differences in bacterial behavior begin to emerge. “The bacteria on the outside are fighting back … to prevent clearance,” says Davis, an assistant professor in Molecular Microbiology and Immunology. “Then there are bacteria that are in the center of this community [that] lack contact with host immune cells, and we became really interested in what was different in the middle relative to the outside.”
To explore those differences, her team devised various fluorescent “reporter” sensors—which light up in response to specific cellular functions or particular molecular signals. Some reporters reveal bacteria-neutrophil binding events, for example, or the presence of nitric oxide, an antimicrobial agent naturally produced by immune cells during infection. This makes it possible to observe which bacteria are directly engaged with the host response against infection based on which microbial cells are glowing and which are not. More recently, Davis’ group has developed a fluorescent protein that allows them to monitor bacterial cell division. This reporter takes longer than other proteins to become active and produce a measurable red glow, which means that only slowly dividing cells will have the opportunity to accumulate enough protein to produce a bright signal.
Importantly, her team applies these reporters in both live rodent models and lab-grown “micro-colonies” that include not just bacteria but neutrophils and other immune cells, providing models of infection that are far more naturalistic than conventional cell culture systems.
Her results to date have illuminated important differences in the behavior of center-dwelling cells versus those on the periphery of bacterial clusters. “The bacteria in the middle seem to be actively dividing while the bacteria on the outside are dealing with the host environment,” she says. “They're modulating the host environment to provide a nicer environment for replication for their relatives in the middle.” Instead of proliferating themselves, the cells on the periphery invest their energy into fending off neutrophils and absorbing and metabolizing incoming nitric oxide.
These community dynamics directly affect antimicrobial drugs’ effectiveness at clearing infections. The cells that enter a quiescent state in order to fend off the immune response are far less susceptible to antibiotics than those that are actively dividing. Such drug “tolerance” can create strongholds of bacteria that persist and regrow after treatment ends. For example, Davis has conducted studies in which animals infected with Yersinia were treated with a standard seven-day clinical course of the antibiotic doxycycline. “We eliminated some of the bacterial cells, but 10% survived,” she says. These survivors are more than sufficient to seed a serious relapsing infection.
It remains to be seen whether other bacterial species exhibit these behaviors, and if other pathogens respond to host immunity and drug treatment in the same fashion. Conlon’s lab principally works with Staphylococcus aureus—one of the more prominent threats in the era of rising antimicrobial resistance—and he observes that “there's common ground there, and some of the mechanisms may well be conserved.” But more research is needed to understand the similarities and differences in how different bacterial species cope with threats to their survival. To this end, some members of Davis’ lab have begun to develop a similar experimental toolbox for S. aureus.
While most of her research has focused on the commonly used antibiotic doxycycline, it’s not yet known whether Yersinia and other bacteria respond similarly to other antibiotics that target distinct biological processes that bacteria rely on to survive and thrive.
Davis’ work is already providing valuable opportunities to develop new drugs or treatment plans that can overcome bacterial drug tolerance and resistance and thus wipe out entire bacterial communities. In some cases, this may simply be a matter of rethinking drug dosing strategies. In one recent collaboration with colleagues at the School of Medicine, for example, Davis helped demonstrate that a shorter course of rifampin treatment with a higher dose was more effective than the current longer, lower-dose regimens at killing off Staphylococcus infection in patients.
Her team has also recently identified a biological pathway in Yersinia cells that could potentially be targeted with drugs to render the microbes more susceptible to doxycycline’s lethal effects. Indeed, multidrug strategies may ultimately prove to be the best solution for dealing with the diverse, cell-specific tactics that bacteria rely on to survive. “We’re going to be utilizing some really cutting-edge technologies to look at individual bacterial cells and see how they're doing things differently,” says Davis.